
Let’s try to get a brief overview of what UMLS is, and then we’ll jump into more details. So, in simple terms, UMLS is a collection of many controlled biomedical vocabularies to provide cross-walk among them and to enable interaction between computer systems. To simplify it further, there are multiple biomedical vocabularies out there developed by different organizations. Each of them has come up with their own set of IDs assigned to each term. UMLS helps in mapping each of them to one common ID.
Let’s take an example of the concept fever. It’s represented by D005334 in Medical Subject Headings (MeSH), 386661006 and 50177009 in SNOMED-CT, 780.6 in ICD-9-CM, 10016558 in the Medical Dictionary for Regulatory Activities terminology (MedDRA), X25 in Perioperative Nursing Data Set, GO:0001660 in Gene Ontology, and U001776 in the Library of Congress Subject Headings. 100 more vocabularies have included fever in their list with a different ID. You can see the problem here: There are as many variations in the IDs as there are vocabularies and there is a need for something that can bring all these terminologies together. That is exactly the purpose of UMLS. It unifies all these codes and maps them to a single biomedical concept (C0015967 in the case of fever).
UMLS was developed by the US National Library of Medicine.
Components to the UMLS knowledge sources
There are three components to the UMLS Knowledge Sources: the Metathesaurus, the Semantic Network, and the SPECIALIST Lexicon and Lexical Tools.
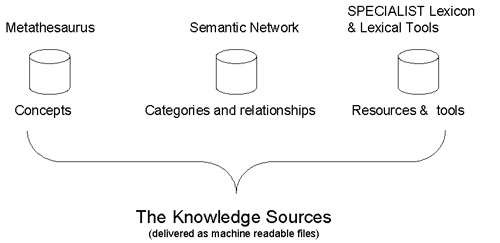
Metathesaurus
Metathesaurus is a key component in the UMLS. It is basically a huge thesaurus created by combining over 200 vocabularies available from various sources. It not only links different vocabularies via a common concept id but also provides relationships between different concepts. For example, it can give you a list of drugs that treats a particular disease or a list of synonyms for a particular drug.
There are over a million concepts in the Metathesaurus and as of April 2021, it has integrated 217 different vocabularies in UMLS. Some examples of vocabularies being MED-RT, RxNorm, MeSH, ICD-10, DrugBank, etc. You can find the complete list here.
The Metathesaurus reflects and preserves the meanings, concept names, and relationships from its source vocabularies. When two different source vocabularies use the same name for differing concepts, the Metathesaurus represents both of the meanings and indicates which meaning is present in which source vocabulary. When conflicting relationships between two concepts appear in different source vocabularies, both views are included in the Metathesaurus.
In other words, the Metathesaurus does not represent a single consistent view of the world. It preserves the many views of the world present in its source vocabularies because these different views may be useful for different tasks. For example, if one vocabulary decides to call fever an annoying little disease, UMLS is not to judge. It’ll show it as it is with the data source mapped to the vocabulary name.
📝 Note:
Metathesaurus is not just limited to drugs and diseases but includes all aspects of the biomedical terms. For example, there are concept ids (CUIs) for Age Groups like “Aged, 80 and over” (C0001795) and “Child” (C0008059) or for Anatomical Structures like “Left set of eyelashes” (C1182718) and “Hand” (C0018563) or for Signs/symptoms like “Abdominal Pain” (C0000737) and “Blushing” (C0005874).
You can read more about Metathesaurus here.
Semantic Network
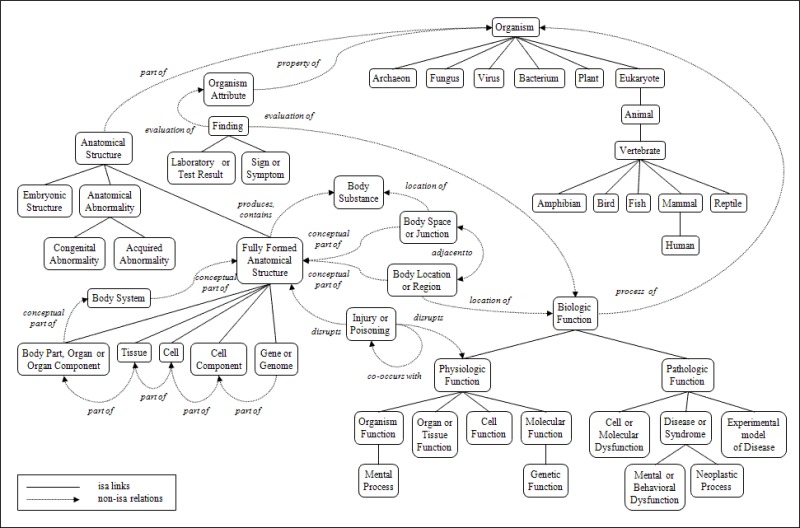
Towards the end of the previous section, notice that we tried to classify the concepts into a few categories. For example, “Abdominal Pain” and “Blushing” are classified as Signs or symptoms while “Aged, 80 and over” and “Child” are classified as Age Groups. Similarly, every concept in the Metathesaurus is assigned at least one such category. These categories are called Semantic Types.
The relationships between Semantic Types are called Semantic Relationships. As an example, take the Semantic Types “Human” and “Mammal”. These are connected by an “isa” relationship (Human is a Mammal). Another example, “Anatomical Structure” is a “part of” “Organism”. This network of relationships between concepts is called Semantic Network.
The purpose of the Semantic Network is to provide a consistent categorization of all concepts represented in the UMLS Metathesaurus and to provide a set of useful relationships between these concepts.
There are total 127 semantic types and 54 relationships in the Semantic Network. You can find a complete list of semantic types here and semantic relationships here.
The 127 Semantic Types are further grouped as:
- Chemical
- Events
- Concepts
- Anatomical structure
- Organisms
- Biological functions
- Physical objects
And the 54 Semantic Relationships are grouped as:
- physically related to
- spatially related to
- temporally related to
- functionally related to
- conceptually related to
The following image shows a Portion of the UMLS Semantic Network Relations.
SPECIALIST Lexicon and Lexical Tools
Before we go ahead let’s quickly revise a few English grammar concepts. Look at these 4 words: prescribe, prescribes, prescribed, and prescription. Each of these words can be used in different contexts but all these words are essentially different word forms for the same expression, that is prescription. In English, such variations are called lexical variations. These variations can also include different spellings for the same expression. For example, anesthetic and anaesthetic are lexical variations of each other.
You may have guessed by now why are we talking about English grammar concepts. Access to biomedical terminologies is often hampered by the high degree of variability inherent in natural language terms and in the terminologies themselves. The SPECIALIST Lexicon and Lexical Tools from UMLS are designed to help users manage this variability.
The SPECIALIST Lexicon is a large syntactic lexicon (vocabulary) of biomedical and general English, which provides the lexical information needed for Natural Language Processing (NLP) systems. Each entry records the syntactic (related to syntax), morphological (a form of words and phrases), and orthographic (like spelling, hyphenation, capitalization, word breaks, and punctuation) information for a term (single word or multiword). Other lexical information for lexical entries, such as derivation pairs and synonym pairs, are generated through systematic methodologies.
The Lexical Tools utilize the Lexicon data to provide a comprehensive toolset and Java application programming interfaces for lexical variant generation and other NLP fundamental functions, including retrieving syntactic category, inflectional variations, spelling variations, abbreviations, acronyms, derivational variations, synonyms, normalization, Unicode-to-ASCII conversion, tokenization, and stop word removal.
Construction of the UMLS Metathesaurus explained with an example
The core idea behind UMLS is that synonymous terms originating from different source vocabularies are clustered into a concept with a preferred term and a Concept Unique Identifier (CUI). The basic building block of the Metathesaurus, also known as an “atom”, is a concept string from each of the source vocabularies. Simply put, each occurrence of a string in each source vocabulary is assigned a unique atom identifier(AUI).
When a lexically identical string appears in multiple source vocabularies for example “Headache” appearing in both MeSH and ICD-10, they are assigned different AUIs. These AUIs are then linked to a single string identifier (SUI) to represent occurrences of the same string. Each SUI is linked to all of its English lexical variants (detected using the Lexical Variant Generator tool) by a common term identifier (LUI). These LUIs may subsequently be linked to more than one CUI due to strings that are lexical variants of each other have different meanings. The following table illustrates how synonymous terms are clustered into a CUI.
| String (Source) | AUI | SUI | LUI | CUI |
|---|---|---|---|---|
| Headache (MeSH) | A0066000 | S0046854 | L0018681 | C0018681 |
| Headache (ICD-10) | A0065992 | |||
| Headaches (MedDRA) | A0066007 | S0046855 | ||
| Headaches (OMIM) | A12003304 | |||
| Cephalodynia (MeSH) | A0540936 | S0475647 | L0380797 |
Source: Construction of UMLS Metathesaurus with Knowledge-Infused Deep Learning
Conclusion
In this blog, we briefly explained the theory behind UMLS. We saw different underlining components that form UMLS and briefly explained each of them. In the next blog, we’ll explore the actual UMLS database and see how a user can find relevant information.
References and Further Reading
- Construction of UMLS Metathesaurus with Knowledge-Infused Deep Learning
- UMLS - Metathesaurus
- UMLS - Semantic Network
- UMLS - SPECIALIST Lexicon and Lexical Tools
- UMLS Semantic Network Fact Sheet
- The Unified Medical Language System SPECIALIST Lexicon and Lexical Tools: Development and applications
- Lexical methods for managing variation in biomedical terminologies
 ChemicBook
ChemicBook
Share it on Twitter Facebook LinkedIn